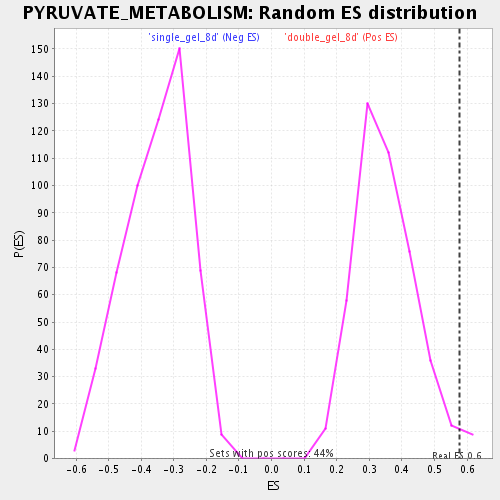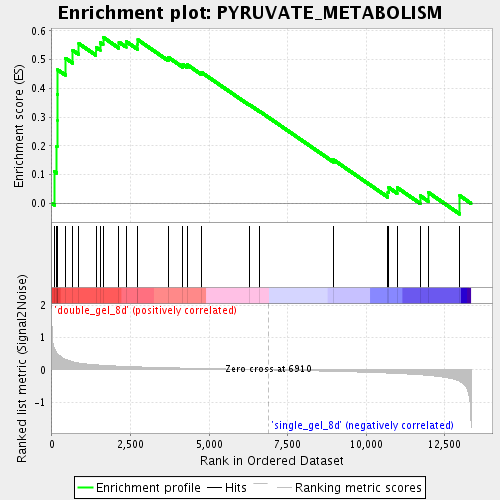
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | PYRUVATE_METABOLISM |
| Enrichment Score (ES) | 0.5777231 |
| Normalized Enrichment Score (NES) | 1.6377788 |
| Nominal p-value | 0.024774775 |
| FDR q-value | 0.053416267 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PC | PC Entrez, Source | pyruvate carboxylase | 79 | 0.652 | 0.1108 | Yes |
| 2 | ACACA | ACACA Entrez, Source | acetyl-Coenzyme A carboxylase alpha | 154 | 0.527 | 0.1996 | Yes |
| 3 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 167 | 0.505 | 0.2891 | Yes |
| 4 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 172 | 0.499 | 0.3781 | Yes |
| 5 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 184 | 0.487 | 0.4644 | Yes |
| 6 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 430 | 0.329 | 0.5050 | Yes |
| 7 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 663 | 0.250 | 0.5322 | Yes |
| 8 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 858 | 0.213 | 0.5557 | Yes |
| 9 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 1403 | 0.157 | 0.5429 | Yes |
| 10 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1535 | 0.147 | 0.5593 | Yes |
| 11 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 1628 | 0.141 | 0.5777 | Yes |
| 12 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 2134 | 0.117 | 0.5607 | No |
| 13 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 2361 | 0.108 | 0.5630 | No |
| 14 | LDHD | LDHD Entrez, Source | lactate dehydrogenase D | 2733 | 0.094 | 0.5521 | No |
| 15 | HAGH | HAGH Entrez, Source | hydroxyacylglutathione hydrolase | 2738 | 0.094 | 0.5686 | No |
| 16 | LDHC | LDHC Entrez, Source | lactate dehydrogenase C | 3707 | 0.066 | 0.5078 | No |
| 17 | MDH2 | MDH2 Entrez, Source | malate dehydrogenase 2, NAD (mitochondrial) | 4168 | 0.056 | 0.4832 | No |
| 18 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 4305 | 0.053 | 0.4824 | No |
| 19 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 4772 | 0.042 | 0.4550 | No |
| 20 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6279 | 0.012 | 0.3441 | No |
| 21 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 6602 | 0.006 | 0.3210 | No |
| 22 | GLO1 | GLO1 Entrez, Source | glyoxalase I | 8965 | -0.045 | 0.1517 | No |
| 23 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 10696 | -0.098 | 0.0393 | No |
| 24 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 10723 | -0.099 | 0.0551 | No |
| 25 | HAGHL | HAGHL Entrez, Source | hydroxyacylglutathione hydrolase-like | 10988 | -0.111 | 0.0551 | No |
| 26 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 11723 | -0.150 | 0.0269 | No |
| 27 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 11983 | -0.171 | 0.0382 | No |
| 28 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 12985 | -0.357 | 0.0268 | No |