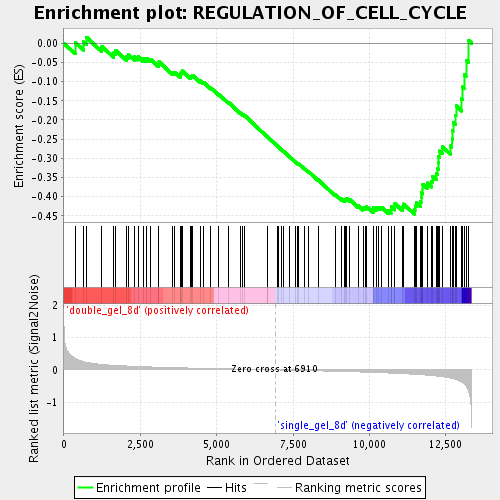
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | REGULATION_OF_CELL_CYCLE |
| Enrichment Score (ES) | -0.4455623 |
| Normalized Enrichment Score (NES) | -1.6097753 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.11260632 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BMP2 | BMP2 Entrez, Source | bone morphogenetic protein 2 | 370 | 0.355 | 0.0022 | No |
| 2 | BMP7 | BMP7 Entrez, Source | bone morphogenetic protein 7 (osteogenic protein 1) | 635 | 0.257 | 0.0041 | No |
| 3 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 740 | 0.230 | 0.0157 | No |
| 4 | PTPRC | PTPRC Entrez, Source | protein tyrosine phosphatase, receptor type, C | 1238 | 0.168 | -0.0076 | No |
| 5 | CDK5R1 | CDK5R1 Entrez, Source | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 1630 | 0.141 | -0.0251 | No |
| 6 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 1695 | 0.137 | -0.0183 | No |
| 7 | ZWINT | ZWINT Entrez, Source | ZW10 interactor | 2055 | 0.119 | -0.0353 | No |
| 8 | BMP4 | BMP4 Entrez, Source | bone morphogenetic protein 4 | 2116 | 0.117 | -0.0299 | No |
| 9 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 2307 | 0.110 | -0.0349 | No |
| 10 | ASNS | ASNS Entrez, Source | asparagine synthetase | 2424 | 0.106 | -0.0347 | No |
| 11 | RAD17 | RAD17 Entrez, Source | RAD17 homolog (S. pombe) | 2601 | 0.099 | -0.0395 | No |
| 12 | RUNX3 | RUNX3 Entrez, Source | runt-related transcription factor 3 | 2707 | 0.095 | -0.0394 | No |
| 13 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 2832 | 0.090 | -0.0411 | No |
| 14 | TBRG4 | TBRG4 Entrez, Source | transforming growth factor beta regulator 4 | 3101 | 0.081 | -0.0544 | No |
| 15 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 3109 | 0.081 | -0.0480 | No |
| 16 | CDK5RAP1 | CDK5RAP1 Entrez, Source | CDK5 regulatory subunit associated protein 1 | 3568 | 0.069 | -0.0767 | No |
| 17 | EREG | EREG Entrez, Source | epiregulin | 3625 | 0.068 | -0.0752 | No |
| 18 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 3805 | 0.064 | -0.0833 | No |
| 19 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 3808 | 0.064 | -0.0780 | No |
| 20 | AIF1 | AIF1 Entrez, Source | allograft inflammatory factor 1 | 3839 | 0.063 | -0.0749 | No |
| 21 | RHOB | RHOB Entrez, Source | ras homolog gene family, member B | 3872 | 0.063 | -0.0720 | No |
| 22 | TBX3 | TBX3 Entrez, Source | T-box 3 (ulnar mammary syndrome) | 4143 | 0.056 | -0.0876 | No |
| 23 | INHA | INHA Entrez, Source | inhibin, alpha | 4183 | 0.055 | -0.0859 | No |
| 24 | PCAF | PCAF Entrez, Source | p300/CBP-associated factor | 4209 | 0.055 | -0.0831 | No |
| 25 | CDK5RAP3 | CDK5RAP3 Entrez, Source | CDK5 regulatory subunit associated protein 3 | 4459 | 0.049 | -0.0977 | No |
| 26 | MAD2L2 | MAD2L2 Entrez, Source | MAD2 mitotic arrest deficient-like 2 (yeast) | 4562 | 0.047 | -0.1014 | No |
| 27 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4801 | 0.042 | -0.1158 | No |
| 28 | GAS7 | GAS7 Entrez, Source | growth arrest-specific 7 | 5073 | 0.036 | -0.1332 | No |
| 29 | NEK6 | NEK6 Entrez, Source | NIMA (never in mitosis gene a)-related kinase 6 | 5393 | 0.030 | -0.1547 | No |
| 30 | BTG4 | BTG4 Entrez, Source | B-cell translocation gene 4 | 5769 | 0.022 | -0.1811 | No |
| 31 | ZBTB17 | ZBTB17 Entrez, Source | zinc finger and BTB domain containing 17 | 5859 | 0.021 | -0.1860 | No |
| 32 | CDC2L5 | CDC2L5 Entrez, Source | cell division cycle 2-like 5 (cholinesterase-related cell division controller) | 5925 | 0.020 | -0.1893 | No |
| 33 | UHMK1 | UHMK1 Entrez, Source | U2AF homology motif (UHM) kinase 1 | 6651 | 0.005 | -0.2436 | No |
| 34 | PLAGL1 | PLAGL1 Entrez, Source | pleiomorphic adenoma gene-like 1 | 6668 | 0.005 | -0.2444 | No |
| 35 | SNF1LK | SNF1LK Entrez, Source | SNF1-like kinase | 6978 | -0.002 | -0.2675 | No |
| 36 | PPM1G | PPM1G Entrez, Source | protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform | 7035 | -0.003 | -0.2715 | No |
| 37 | SERTAD1 | SERTAD1 Entrez, Source | SERTA domain containing 1 | 7125 | -0.005 | -0.2779 | No |
| 38 | MFN2 | MFN2 Entrez, Source | mitofusin 2 | 7196 | -0.006 | -0.2826 | No |
| 39 | INHBA | INHBA Entrez, Source | inhibin, beta A (activin A, activin AB alpha polypeptide) | 7375 | -0.010 | -0.2952 | No |
| 40 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 7589 | -0.014 | -0.3101 | No |
| 41 | GTPBP4 | GTPBP4 Entrez, Source | GTP binding protein 4 | 7656 | -0.016 | -0.3138 | No |
| 42 | MAP2K6 | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 7666 | -0.016 | -0.3131 | No |
| 43 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 7859 | -0.020 | -0.3259 | No |
| 44 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 8008 | -0.024 | -0.3351 | No |
| 45 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 8322 | -0.030 | -0.3561 | No |
| 46 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 8877 | -0.043 | -0.3943 | No |
| 47 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9083 | -0.048 | -0.4057 | No |
| 48 | NPM2 | NPM2 Entrez, Source | nucleophosmin/nucleoplasmin, 2 | 9187 | -0.051 | -0.4091 | No |
| 49 | CUL5 | CUL5 Entrez, Source | cullin 5 | 9220 | -0.052 | -0.4071 | No |
| 50 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 9252 | -0.053 | -0.4050 | No |
| 51 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 9345 | -0.056 | -0.4072 | No |
| 52 | ZW10 | ZW10 Entrez, Source | ZW10, kinetochore associated, homolog (Drosophila) | 9627 | -0.063 | -0.4230 | No |
| 53 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 9788 | -0.068 | -0.4294 | No |
| 54 | PPP1R9B | PPP1R9B Entrez, Source | protein phosphatase 1, regulatory subunit 9B, spinophilin | 9861 | -0.070 | -0.4289 | No |
| 55 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 9912 | -0.072 | -0.4265 | No |
| 56 | NBN | NBN Entrez, Source | nibrin | 10122 | -0.079 | -0.4356 | No |
| 57 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 10127 | -0.079 | -0.4292 | No |
| 58 | APBB1 | APBB1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | 10220 | -0.082 | -0.4292 | No |
| 59 | APC | APC Entrez, Source | adenomatosis polyposis coli | 10296 | -0.085 | -0.4277 | No |
| 60 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 10391 | -0.088 | -0.4273 | No |
| 61 | RASSF1 | RASSF1 Entrez, Source | Ras association (RalGDS/AF-6) domain family 1 | 10613 | -0.096 | -0.4359 | Yes |
| 62 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 10707 | -0.099 | -0.4345 | Yes |
| 63 | CHFR | CHFR Entrez, Source | checkpoint with forkhead and ring finger domains | 10712 | -0.099 | -0.4264 | Yes |
| 64 | MADD | MADD Entrez, Source | MAP-kinase activating death domain | 10825 | -0.104 | -0.4261 | Yes |
| 65 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 10832 | -0.104 | -0.4177 | Yes |
| 66 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 11087 | -0.116 | -0.4270 | Yes |
| 67 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 11117 | -0.118 | -0.4192 | Yes |
| 68 | CDC16 | CDC16 Entrez, Source | CDC16 cell division cycle 16 homolog (S. cerevisiae) | 11467 | -0.135 | -0.4341 | Yes |
| 69 | CDK10 | CDK10 Entrez, Source | cyclin-dependent kinase (CDC2-like) 10 | 11493 | -0.137 | -0.4244 | Yes |
| 70 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 11527 | -0.139 | -0.4151 | Yes |
| 71 | BTG3 | BTG3 Entrez, Source | BTG family, member 3 | 11661 | -0.148 | -0.4126 | Yes |
| 72 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 11690 | -0.149 | -0.4020 | Yes |
| 73 | TBRG1 | TBRG1 Entrez, Source | transforming growth factor beta regulator 1 | 11701 | -0.150 | -0.3901 | Yes |
| 74 | CCNG1 | CCNG1 Entrez, Source | cyclin G1 | 11744 | -0.152 | -0.3804 | Yes |
| 75 | CD28 | CD28 Entrez, Source | CD28 molecule | 11745 | -0.152 | -0.3675 | Yes |
| 76 | DDB1 | DDB1 Entrez, Source | damage-specific DNA binding protein 1, 127kDa | 11887 | -0.163 | -0.3643 | Yes |
| 77 | CHEK2 | CHEK2 Entrez, Source | CHK2 checkpoint homolog (S. pombe) | 12031 | -0.176 | -0.3602 | Yes |
| 78 | KHDRBS1 | KHDRBS1 Entrez, Source | KH domain containing, RNA binding, signal transduction associated 1 | 12056 | -0.178 | -0.3469 | Yes |
| 79 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 12182 | -0.190 | -0.3402 | Yes |
| 80 | MAPK12 | MAPK12 Entrez, Source | mitogen-activated protein kinase 12 | 12219 | -0.194 | -0.3265 | Yes |
| 81 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 12251 | -0.199 | -0.3120 | Yes |
| 82 | ANAPC2 | ANAPC2 Entrez, Source | anaphase promoting complex subunit 2 | 12258 | -0.199 | -0.2955 | Yes |
| 83 | CDC37 | CDC37 Entrez, Source | CDC37 cell division cycle 37 homolog (S. cerevisiae) | 12294 | -0.204 | -0.2809 | Yes |
| 84 | ING4 | ING4 Entrez, Source | inhibitor of growth family, member 4 | 12380 | -0.212 | -0.2693 | Yes |
| 85 | TIMELESS | TIMELESS Entrez, Source | timeless homolog (Drosophila) | 12644 | -0.253 | -0.2677 | Yes |
| 86 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 12700 | -0.264 | -0.2494 | Yes |
| 87 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 12725 | -0.270 | -0.2283 | Yes |
| 88 | NOTCH2 | NOTCH2 Entrez, Source | Notch homolog 2 (Drosophila) | 12734 | -0.272 | -0.2058 | Yes |
| 89 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12821 | -0.294 | -0.1874 | Yes |
| 90 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12832 | -0.296 | -0.1630 | Yes |
| 91 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 13016 | -0.374 | -0.1451 | Yes |
| 92 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 13047 | -0.395 | -0.1139 | Yes |
| 93 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 13099 | -0.425 | -0.0817 | Yes |
| 94 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13162 | -0.500 | -0.0439 | Yes |
| 95 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13247 | -0.677 | 0.0072 | Yes |