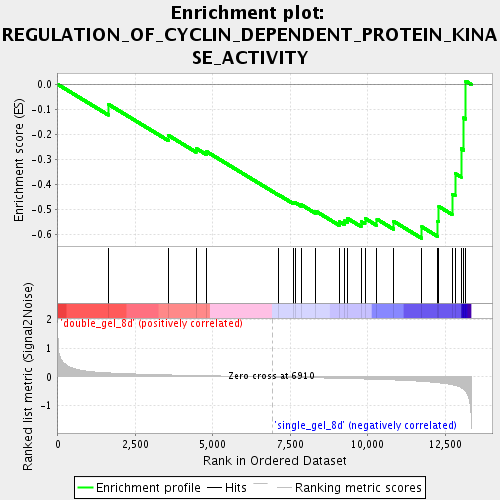
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | REGULATION_OF_CYCLIN_DEPENDENT_PROTEIN_KINASE_ACTIVITY |
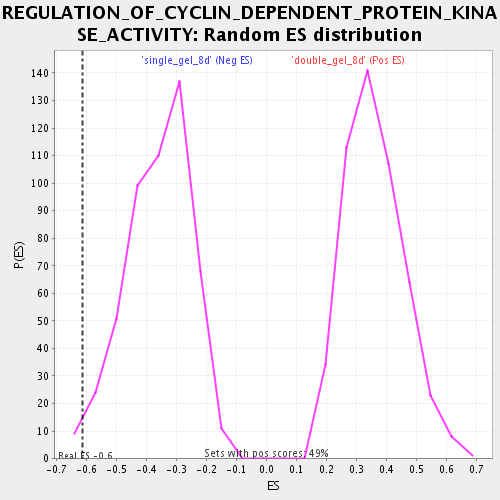
| Enrichment Score (ES) | -0.61559445 |
| Normalized Enrichment Score (NES) | -1.6943994 |
| Nominal p-value | 0.015717093 |
| FDR q-value | 0.07773607 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDK5R1 | CDK5R1 Entrez, Source | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 1630 | 0.141 | -0.0794 | No |
| 2 | CDK5RAP1 | CDK5RAP1 Entrez, Source | CDK5 regulatory subunit associated protein 1 | 3568 | 0.069 | -0.2038 | No |
| 3 | CDK5RAP3 | CDK5RAP3 Entrez, Source | CDK5 regulatory subunit associated protein 3 | 4459 | 0.049 | -0.2557 | No |
| 4 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4801 | 0.042 | -0.2686 | No |
| 5 | SERTAD1 | SERTAD1 Entrez, Source | SERTA domain containing 1 | 7125 | -0.005 | -0.4416 | No |
| 6 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 7589 | -0.014 | -0.4720 | No |
| 7 | GTPBP4 | GTPBP4 Entrez, Source | GTP binding protein 4 | 7656 | -0.016 | -0.4722 | No |
| 8 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 7859 | -0.020 | -0.4813 | No |
| 9 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 8322 | -0.030 | -0.5067 | No |
| 10 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9083 | -0.048 | -0.5492 | No |
| 11 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 9252 | -0.053 | -0.5457 | No |
| 12 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 9345 | -0.056 | -0.5357 | No |
| 13 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 9788 | -0.068 | -0.5483 | No |
| 14 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 9912 | -0.072 | -0.5357 | No |
| 15 | APC | APC Entrez, Source | adenomatosis polyposis coli | 10296 | -0.085 | -0.5387 | No |
| 16 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 10832 | -0.104 | -0.5472 | No |
| 17 | CCNG1 | CCNG1 Entrez, Source | cyclin G1 | 11744 | -0.152 | -0.5695 | Yes |
| 18 | ANAPC2 | ANAPC2 Entrez, Source | anaphase promoting complex subunit 2 | 12258 | -0.199 | -0.5473 | Yes |
| 19 | CDC37 | CDC37 Entrez, Source | CDC37 cell division cycle 37 homolog (S. cerevisiae) | 12294 | -0.204 | -0.4880 | Yes |
| 20 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 12725 | -0.270 | -0.4382 | Yes |
| 21 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12821 | -0.294 | -0.3560 | Yes |
| 22 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 13016 | -0.374 | -0.2568 | Yes |
| 23 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 13099 | -0.425 | -0.1338 | Yes |
| 24 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13162 | -0.500 | 0.0135 | Yes |