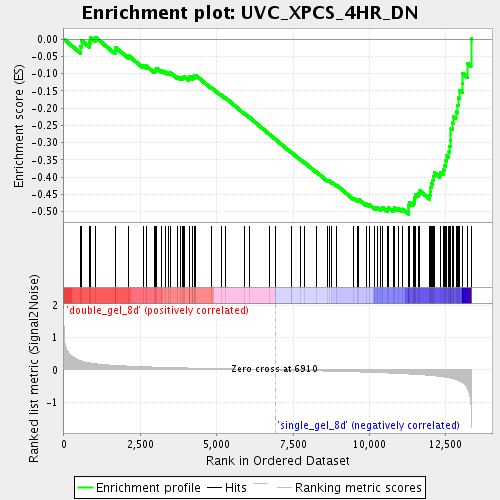
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | UVC_XPCS_4HR_DN |
| Enrichment Score (ES) | -0.50682116 |
| Normalized Enrichment Score (NES) | -1.8332322 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03825836 |
| FWER p-Value | 0.792 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ACSL3 | ACSL3 Entrez, Source | acyl-CoA synthetase long-chain family member 3 | 546 | 0.284 | -0.0210 | No |
| 2 | BHLHB2 | BHLHB2 Entrez, Source | basic helix-loop-helix domain containing, class B, 2 | 581 | 0.272 | -0.0041 | No |
| 3 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 837 | 0.215 | -0.0080 | No |
| 4 | ZFHX1B | ZFHX1B Entrez, Source | zinc finger homeobox 1b | 883 | 0.209 | 0.0036 | No |
| 5 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 1037 | 0.188 | 0.0055 | No |
| 6 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 1676 | 0.138 | -0.0328 | No |
| 7 | FYN | FYN Entrez, Source | FYN oncogene related to SRC, FGR, YES | 1691 | 0.137 | -0.0241 | No |
| 8 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 2102 | 0.118 | -0.0466 | No |
| 9 | TGFBR3 | TGFBR3 Entrez, Source | transforming growth factor, beta receptor III (betaglycan, 300kDa) | 2590 | 0.100 | -0.0763 | No |
| 10 | PRR3 | PRR3 Entrez, Source | proline rich 3 | 2697 | 0.096 | -0.0775 | No |
| 11 | MECP2 | MECP2 Entrez, Source | methyl CpG binding protein 2 (Rett syndrome) | 2966 | 0.086 | -0.0916 | No |
| 12 | RGS7 | RGS7 Entrez, Source | regulator of G-protein signalling 7 | 3011 | 0.085 | -0.0889 | No |
| 13 | DLC1 | DLC1 Entrez, Source | deleted in liver cancer 1 | 3032 | 0.084 | -0.0844 | No |
| 14 | BTRC | BTRC Entrez, Source | beta-transducin repeat containing | 3196 | 0.079 | -0.0910 | No |
| 15 | IL6 | IL6 Entrez, Source | interleukin 6 (interferon, beta 2) | 3320 | 0.076 | -0.0949 | No |
| 16 | MARK3 | MARK3 Entrez, Source | MAP/microtubule affinity-regulating kinase 3 | 3431 | 0.073 | -0.0980 | No |
| 17 | TERF1 | TERF1 Entrez, Source | telomeric repeat binding factor (NIMA-interacting) 1 | 3490 | 0.071 | -0.0973 | No |
| 18 | CEP350 | CEP350 Entrez, Source | centrosomal protein 350kDa | 3715 | 0.066 | -0.1095 | No |
| 19 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 3805 | 0.064 | -0.1117 | No |
| 20 | ZHX3 | ZHX3 Entrez, Source | zinc fingers and homeoboxes 3 | 3874 | 0.063 | -0.1123 | No |
| 21 | CREB1 | CREB1 Entrez, Source | cAMP responsive element binding protein 1 | 3915 | 0.062 | -0.1109 | No |
| 22 | FGF5 | FGF5 Entrez, Source | fibroblast growth factor 5 | 3936 | 0.061 | -0.1081 | No |
| 23 | USP15 | USP15 Entrez, Source | ubiquitin specific peptidase 15 | 4093 | 0.057 | -0.1158 | No |
| 24 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 4096 | 0.057 | -0.1118 | No |
| 25 | IGF1R | IGF1R Entrez, Source | insulin-like growth factor 1 receptor | 4105 | 0.057 | -0.1084 | No |
| 26 | SOS2 | SOS2 Entrez, Source | son of sevenless homolog 2 (Drosophila) | 4217 | 0.055 | -0.1128 | No |
| 27 | CNOT4 | CNOT4 Entrez, Source | CCR4-NOT transcription complex, subunit 4 | 4218 | 0.055 | -0.1089 | No |
| 28 | BMPR1A | BMPR1A Entrez, Source | bone morphogenetic protein receptor, type IA | 4277 | 0.053 | -0.1095 | No |
| 29 | GLRB | GLRB Entrez, Source | glycine receptor, beta | 4280 | 0.053 | -0.1058 | No |
| 30 | CENTB2 | CENTB2 Entrez, Source | centaurin, beta 2 | 4313 | 0.052 | -0.1045 | No |
| 31 | KLF6 | KLF6 Entrez, Source | Kruppel-like factor 6 | 4836 | 0.041 | -0.1410 | No |
| 32 | APPBP2 | APPBP2 Entrez, Source | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 5141 | 0.035 | -0.1614 | No |
| 33 | BARD1 | BARD1 Entrez, Source | BRCA1 associated RING domain 1 | 5278 | 0.032 | -0.1694 | No |
| 34 | CDC2L5 | CDC2L5 Entrez, Source | cell division cycle 2-like 5 (cholinesterase-related cell division controller) | 5925 | 0.020 | -0.2168 | No |
| 35 | PTPN12 | PTPN12 Entrez, Source | protein tyrosine phosphatase, non-receptor type 12 | 6058 | 0.017 | -0.2255 | No |
| 36 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 6727 | 0.004 | -0.2757 | No |
| 37 | CUTL1 | CUTL1 Entrez, Source | cut-like 1, CCAAT displacement protein (Drosophila) | 6916 | -0.000 | -0.2899 | No |
| 38 | ATP2B1 | ATP2B1 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 1 | 6925 | -0.000 | -0.2905 | No |
| 39 | TRIM2 | TRIM2 Entrez, Source | tripartite motif-containing 2 | 7437 | -0.011 | -0.3282 | No |
| 40 | AFAP | AFAP Entrez, Source | - | 7458 | -0.011 | -0.3289 | No |
| 41 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 7740 | -0.017 | -0.3489 | No |
| 42 | EIF2C2 | EIF2C2 Entrez, Source | eukaryotic translation initiation factor 2C, 2 | 7880 | -0.021 | -0.3580 | No |
| 43 | NIPBL | NIPBL Entrez, Source | Nipped-B homolog (Drosophila) | 8250 | -0.029 | -0.3838 | No |
| 44 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 8626 | -0.037 | -0.4094 | No |
| 45 | TIAM1 | TIAM1 Entrez, Source | T-cell lymphoma invasion and metastasis 1 | 8679 | -0.039 | -0.4106 | No |
| 46 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 8768 | -0.040 | -0.4144 | No |
| 47 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 8920 | -0.044 | -0.4226 | No |
| 48 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 9477 | -0.059 | -0.4604 | No |
| 49 | ARHGEF7 | ARHGEF7 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 7 | 9623 | -0.063 | -0.4668 | No |
| 50 | GNAQ | GNAQ Entrez, Source | guanine nucleotide binding protein (G protein), q polypeptide | 9655 | -0.064 | -0.4646 | No |
| 51 | STRN3 | STRN3 Entrez, Source | striatin, calmodulin binding protein 3 | 9894 | -0.071 | -0.4774 | No |
| 52 | RSN | RSN Entrez, Source | restin (Reed-Steinberg cell-expressed intermediate filament-associated protein) | 9993 | -0.074 | -0.4795 | No |
| 53 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 10171 | -0.080 | -0.4872 | No |
| 54 | RBM16 | RBM16 Entrez, Source | RNA binding motif protein 16 | 10272 | -0.084 | -0.4887 | No |
| 55 | NCOA3 | NCOA3 Entrez, Source | nuclear receptor coactivator 3 | 10363 | -0.087 | -0.4893 | No |
| 56 | CNOT2 | CNOT2 Entrez, Source | CCR4-NOT transcription complex, subunit 2 | 10411 | -0.089 | -0.4865 | No |
| 57 | RBM39 | RBM39 Entrez, Source | RNA binding motif protein 39 | 10593 | -0.095 | -0.4934 | No |
| 58 | PHLPP | PHLPP Entrez, Source | PH domain and leucine rich repeat protein phosphatase | 10612 | -0.095 | -0.4880 | No |
| 59 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 10778 | -0.102 | -0.4931 | No |
| 60 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 10819 | -0.104 | -0.4888 | No |
| 61 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 10938 | -0.109 | -0.4899 | No |
| 62 | PLEKHC1 | PLEKHC1 Entrez, Source | pleckstrin homology domain containing, family C (with FERM domain) member 1 | 11076 | -0.115 | -0.4920 | No |
| 63 | AKAP9 | AKAP9 Entrez, Source | A kinase (PRKA) anchor protein (yotiao) 9 | 11273 | -0.125 | -0.4979 | Yes |
| 64 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 11278 | -0.125 | -0.4892 | Yes |
| 65 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 11280 | -0.126 | -0.4803 | Yes |
| 66 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 11301 | -0.126 | -0.4728 | Yes |
| 67 | STK3 | STK3 Entrez, Source | serine/threonine kinase 3 (STE20 homolog, yeast) | 11428 | -0.133 | -0.4729 | Yes |
| 68 | E2F5 | E2F5 Entrez, Source | E2F transcription factor 5, p130-binding | 11463 | -0.135 | -0.4658 | Yes |
| 69 | TUBGCP3 | TUBGCP3 Entrez, Source | tubulin, gamma complex associated protein 3 | 11484 | -0.136 | -0.4576 | Yes |
| 70 | MARCH7 | MARCH7 Entrez, Source | membrane-associated ring finger (C3HC4) 7 | 11511 | -0.138 | -0.4497 | Yes |
| 71 | ATF2 | ATF2 Entrez, Source | activating transcription factor 2 | 11597 | -0.143 | -0.4459 | Yes |
| 72 | WNT5A | WNT5A Entrez, Source | wingless-type MMTV integration site family, member 5A | 11650 | -0.147 | -0.4393 | Yes |
| 73 | EDD1 | EDD1 Entrez, Source | E3 ubiquitin protein ligase, HECT domain containing, 1 | 11974 | -0.171 | -0.4515 | Yes |
| 74 | PUM2 | PUM2 Entrez, Source | pumilio homolog 2 (Drosophila) | 11991 | -0.172 | -0.4404 | Yes |
| 75 | ACVR1 | ACVR1 Entrez, Source | activin A receptor, type I | 11998 | -0.173 | -0.4285 | Yes |
| 76 | PARG | PARG Entrez, Source | poly (ADP-ribose) glycohydrolase | 12022 | -0.175 | -0.4178 | Yes |
| 77 | NFIB | NFIB Entrez, Source | nuclear factor I/B | 12064 | -0.179 | -0.4081 | Yes |
| 78 | TLE4 | TLE4 Entrez, Source | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) | 12093 | -0.181 | -0.3973 | Yes |
| 79 | TSPAN5 | TSPAN5 Entrez, Source | tetraspanin 5 | 12120 | -0.183 | -0.3861 | Yes |
| 80 | MTMR3 | MTMR3 Entrez, Source | myotubularin related protein 3 | 12309 | -0.205 | -0.3857 | Yes |
| 81 | PTK2 | PTK2 Entrez, Source | PTK2 protein tyrosine kinase 2 | 12413 | -0.216 | -0.3780 | Yes |
| 82 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 12463 | -0.223 | -0.3658 | Yes |
| 83 | REV3L | REV3L Entrez, Source | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) | 12493 | -0.227 | -0.3518 | Yes |
| 84 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 12533 | -0.233 | -0.3381 | Yes |
| 85 | TRIO | TRIO Entrez, Source | triple functional domain (PTPRF interacting) | 12599 | -0.245 | -0.3255 | Yes |
| 86 | KIF2A | KIF2A Entrez, Source | kinesin heavy chain member 2A | 12626 | -0.249 | -0.3096 | Yes |
| 87 | PTPRK | PTPRK Entrez, Source | protein tyrosine phosphatase, receptor type, K | 12652 | -0.254 | -0.2934 | Yes |
| 88 | RRAS2 | RRAS2 Entrez, Source | related RAS viral (r-ras) oncogene homolog 2 | 12665 | -0.257 | -0.2760 | Yes |
| 89 | MTF2 | MTF2 Entrez, Source | metal response element binding transcription factor 2 | 12667 | -0.258 | -0.2577 | Yes |
| 90 | UGCG | UGCG Entrez, Source | UDP-glucose ceramide glucosyltransferase | 12723 | -0.270 | -0.2426 | Yes |
| 91 | CEP170 | CEP170 Entrez, Source | centrosomal protein 170kDa | 12737 | -0.273 | -0.2240 | Yes |
| 92 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12832 | -0.296 | -0.2100 | Yes |
| 93 | PCTK2 | PCTK2 Entrez, Source | PCTAIRE protein kinase 2 | 12886 | -0.316 | -0.1915 | Yes |
| 94 | SCHIP1 | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 12910 | -0.326 | -0.1699 | Yes |
| 95 | RUNX1 | RUNX1 Entrez, Source | runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 12949 | -0.338 | -0.1487 | Yes |
| 96 | ZNF292 | ZNF292 Entrez, Source | zinc finger protein 292 | 13045 | -0.393 | -0.1278 | Yes |
| 97 | GNAI1 | GNAI1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 13055 | -0.401 | -0.0998 | Yes |
| 98 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 13212 | -0.580 | -0.0701 | Yes |
| 99 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 13324 | -1.119 | 0.0014 | Yes |