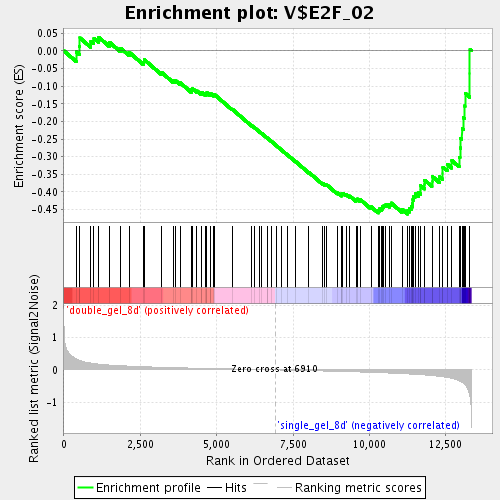
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | single_gel_8d |
| GeneSet | V$E2F_02 |
| Enrichment Score (ES) | -0.46275803 |
| Normalized Enrichment Score (NES) | -1.6492692 |
| Nominal p-value | 0.0053859963 |
| FDR q-value | 0.096968494 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FKBP5 | FKBP5 Entrez, Source | FK506 binding protein 5 | 405 | 0.337 | -0.0028 | No |
| 2 | ATF5 | ATF5 Entrez, Source | activating transcription factor 5 | 511 | 0.296 | 0.0136 | No |
| 3 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 524 | 0.292 | 0.0367 | No |
| 4 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 880 | 0.210 | 0.0271 | No |
| 5 | MRPL40 | MRPL40 Entrez, Source | mitochondrial ribosomal protein L40 | 981 | 0.197 | 0.0358 | No |
| 6 | NELL2 | NELL2 Entrez, Source | NEL-like 2 (chicken) | 1138 | 0.178 | 0.0386 | No |
| 7 | EFNA5 | EFNA5 Entrez, Source | ephrin-A5 | 1498 | 0.150 | 0.0238 | No |
| 8 | PCSK1 | PCSK1 Entrez, Source | proprotein convertase subtilisin/kexin type 1 | 1853 | 0.129 | 0.0077 | No |
| 9 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 2146 | 0.116 | -0.0048 | No |
| 10 | SLC9A5 | SLC9A5 Entrez, Source | solute carrier family 9 (sodium/hydrogen exchanger), member 5 | 2614 | 0.099 | -0.0319 | No |
| 11 | USP52 | USP52 Entrez, Source | ubiquitin specific peptidase 52 | 2621 | 0.098 | -0.0243 | No |
| 12 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 3194 | 0.079 | -0.0610 | No |
| 13 | CORT | CORT Entrez, Source | cortistatin | 3589 | 0.069 | -0.0850 | No |
| 14 | KCNA6 | KCNA6 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 6 | 3661 | 0.067 | -0.0848 | No |
| 15 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 3805 | 0.064 | -0.0904 | No |
| 16 | PHF5A | PHF5A Entrez, Source | PHD finger protein 5A | 4179 | 0.055 | -0.1140 | No |
| 17 | HTF9C | HTF9C Entrez, Source | - | 4181 | 0.055 | -0.1095 | No |
| 18 | GATA1 | GATA1 Entrez, Source | GATA binding protein 1 (globin transcription factor 1) | 4211 | 0.055 | -0.1072 | No |
| 19 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 4353 | 0.052 | -0.1136 | No |
| 20 | FANCC | FANCC Entrez, Source | Fanconi anemia, complementation group C | 4494 | 0.049 | -0.1201 | No |
| 21 | GRIA4 | GRIA4 Entrez, Source | glutamate receptor, ionotrophic, AMPA 4 | 4512 | 0.048 | -0.1175 | No |
| 22 | POLD3 | POLD3 Entrez, Source | polymerase (DNA-directed), delta 3, accessory subunit | 4626 | 0.046 | -0.1222 | No |
| 23 | SP3 | SP3 Entrez, Source | Sp3 transcription factor | 4669 | 0.045 | -0.1217 | No |
| 24 | KCNS2 | KCNS2 Entrez, Source | potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 | 4679 | 0.044 | -0.1188 | No |
| 25 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 4801 | 0.042 | -0.1244 | No |
| 26 | MAPT | MAPT Entrez, Source | microtubule-associated protein tau | 4812 | 0.042 | -0.1218 | No |
| 27 | GABRB3 | GABRB3 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, beta 3 | 4900 | 0.040 | -0.1251 | No |
| 28 | MXD3 | MXD3 Entrez, Source | MAX dimerization protein 3 | 4925 | 0.039 | -0.1236 | No |
| 29 | KBTBD7 | KBTBD7 Entrez, Source | kelch repeat and BTB (POZ) domain containing 7 | 5504 | 0.028 | -0.1650 | No |
| 30 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 6146 | 0.015 | -0.2121 | No |
| 31 | PRPF4B | PRPF4B Entrez, Source | PRP4 pre-mRNA processing factor 4 homolog B (yeast) | 6222 | 0.014 | -0.2166 | No |
| 32 | UGCGL1 | UGCGL1 Entrez, Source | UDP-glucose ceramide glucosyltransferase-like 1 | 6411 | 0.009 | -0.2300 | No |
| 33 | GLRA3 | GLRA3 Entrez, Source | glycine receptor, alpha 3 | 6466 | 0.008 | -0.2334 | No |
| 34 | PAQR4 | PAQR4 Entrez, Source | progestin and adipoQ receptor family member IV | 6650 | 0.005 | -0.2468 | No |
| 35 | PLAGL1 | PLAGL1 Entrez, Source | pleiomorphic adenoma gene-like 1 | 6668 | 0.005 | -0.2477 | No |
| 36 | TRIM39 | TRIM39 Entrez, Source | tripartite motif-containing 39 | 6792 | 0.003 | -0.2567 | No |
| 37 | ACBD6 | ACBD6 Entrez, Source | acyl-Coenzyme A binding domain containing 6 | 6961 | -0.001 | -0.2693 | No |
| 38 | MAPK6 | MAPK6 Entrez, Source | mitogen-activated protein kinase 6 | 7114 | -0.004 | -0.2804 | No |
| 39 | SUMO1 | SUMO1 Entrez, Source | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | 7316 | -0.009 | -0.2949 | No |
| 40 | HCN3 | HCN3 Entrez, Source | hyperpolarization activated cyclic nucleotide-gated potassium channel 3 | 7586 | -0.014 | -0.3140 | No |
| 41 | RPS20 | RPS20 Entrez, Source | ribosomal protein S20 | 8019 | -0.024 | -0.3447 | No |
| 42 | SFRS7 | SFRS7 Entrez, Source | splicing factor, arginine/serine-rich 7, 35kDa | 8464 | -0.033 | -0.3754 | No |
| 43 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 8540 | -0.035 | -0.3782 | No |
| 44 | POU4F1 | POU4F1 Entrez, Source | POU domain, class 4, transcription factor 1 | 8602 | -0.037 | -0.3798 | No |
| 45 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 8949 | -0.045 | -0.4022 | No |
| 46 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 9075 | -0.048 | -0.4077 | No |
| 47 | NCL | NCL Entrez, Source | nucleolin | 9096 | -0.049 | -0.4052 | No |
| 48 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 9128 | -0.050 | -0.4034 | No |
| 49 | FMO4 | FMO4 Entrez, Source | flavin containing monooxygenase 4 | 9231 | -0.052 | -0.4069 | No |
| 50 | HOXC10 | HOXC10 Entrez, Source | homeobox C10 | 9351 | -0.056 | -0.4112 | No |
| 51 | GSPT1 | GSPT1 Entrez, Source | G1 to S phase transition 1 | 9560 | -0.061 | -0.4219 | No |
| 52 | RQCD1 | RQCD1 Entrez, Source | RCD1 required for cell differentiation1 homolog (S. pombe) | 9601 | -0.063 | -0.4198 | No |
| 53 | SFRS10 | SFRS10 Entrez, Source | splicing factor, arginine/serine-rich 10 (transformer 2 homolog, Drosophila) | 9695 | -0.065 | -0.4214 | No |
| 54 | PAX6 | PAX6 Entrez, Source | paired box gene 6 (aniridia, keratitis) | 10052 | -0.076 | -0.4420 | No |
| 55 | ING3 | ING3 Entrez, Source | inhibitor of growth family, member 3 | 10287 | -0.084 | -0.4528 | Yes |
| 56 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 10317 | -0.085 | -0.4479 | Yes |
| 57 | XTP3TPA | XTP3TPA Entrez, Source | - | 10388 | -0.088 | -0.4460 | Yes |
| 58 | EIF4A1 | EIF4A1 Entrez, Source | eukaryotic translation initiation factor 4A, isoform 1 | 10424 | -0.089 | -0.4413 | Yes |
| 59 | TYRO3 | TYRO3 Entrez, Source | TYRO3 protein tyrosine kinase | 10473 | -0.091 | -0.4375 | Yes |
| 60 | DNMT1 | DNMT1 Entrez, Source | DNA (cytosine-5-)-methyltransferase 1 | 10538 | -0.093 | -0.4347 | Yes |
| 61 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 10651 | -0.097 | -0.4351 | Yes |
| 62 | RET | RET Entrez, Source | ret proto-oncogene (multiple endocrine neoplasia and medullary thyroid carcinoma 1, Hirschsprung disease) | 10708 | -0.099 | -0.4312 | Yes |
| 63 | CDC5L | CDC5L Entrez, Source | CDC5 cell division cycle 5-like (S. pombe) | 11068 | -0.115 | -0.4489 | Yes |
| 64 | CDCA7 | CDCA7 Entrez, Source | cell division cycle associated 7 | 11253 | -0.124 | -0.4525 | Yes |
| 65 | HNRPR | HNRPR Entrez, Source | heterogeneous nuclear ribonucleoprotein R | 11314 | -0.127 | -0.4466 | Yes |
| 66 | NUP62 | NUP62 Entrez, Source | nucleoporin 62kDa | 11369 | -0.129 | -0.4400 | Yes |
| 67 | DCK | DCK Entrez, Source | deoxycytidine kinase | 11406 | -0.131 | -0.4320 | Yes |
| 68 | CSPG2 | CSPG2 Entrez, Source | chondroitin sulfate proteoglycan 2 (versican) | 11420 | -0.132 | -0.4221 | Yes |
| 69 | ILF3 | ILF3 Entrez, Source | interleukin enhancer binding factor 3, 90kDa | 11444 | -0.134 | -0.4128 | Yes |
| 70 | TLE3 | TLE3 Entrez, Source | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | 11492 | -0.137 | -0.4050 | Yes |
| 71 | MYH10 | MYH10 Entrez, Source | myosin, heavy chain 10, non-muscle | 11608 | -0.144 | -0.4019 | Yes |
| 72 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 11672 | -0.148 | -0.3944 | Yes |
| 73 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 11673 | -0.148 | -0.3823 | Yes |
| 74 | FANCD2 | FANCD2 Entrez, Source | Fanconi anemia, complementation group D2 | 11799 | -0.156 | -0.3788 | Yes |
| 75 | CBX3 | CBX3 Entrez, Source | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | 11802 | -0.156 | -0.3661 | Yes |
| 76 | HMGA1 | HMGA1 Entrez, Source | high mobility group AT-hook 1 | 12047 | -0.177 | -0.3700 | Yes |
| 77 | SMAD6 | SMAD6 Entrez, Source | SMAD, mothers against DPP homolog 6 (Drosophila) | 12050 | -0.177 | -0.3555 | Yes |
| 78 | PPIG | PPIG Entrez, Source | peptidylprolyl isomerase G (cyclophilin G) | 12293 | -0.204 | -0.3570 | Yes |
| 79 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 12401 | -0.215 | -0.3474 | Yes |
| 80 | TMPO | TMPO Entrez, Source | thymopoietin | 12402 | -0.215 | -0.3298 | Yes |
| 81 | EPHB1 | EPHB1 Entrez, Source | EPH receptor B1 | 12545 | -0.235 | -0.3212 | Yes |
| 82 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 12695 | -0.263 | -0.3108 | Yes |
| 83 | PRPS2 | PRPS2 Entrez, Source | phosphoribosyl pyrophosphate synthetase 2 | 12939 | -0.335 | -0.3016 | Yes |
| 84 | SLCO3A1 | SLCO3A1 Entrez, Source | solute carrier organic anion transporter family, member 3A1 | 12964 | -0.345 | -0.2750 | Yes |
| 85 | SOAT1 | SOAT1 Entrez, Source | sterol O-acyltransferase (acyl-Coenzyme A: cholesterol acyltransferase) 1 | 12979 | -0.351 | -0.2472 | Yes |
| 86 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13028 | -0.381 | -0.2196 | Yes |
| 87 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 13077 | -0.411 | -0.1894 | Yes |
| 88 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 13120 | -0.443 | -0.1561 | Yes |
| 89 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 13128 | -0.448 | -0.1198 | Yes |
| 90 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 13288 | -0.823 | -0.0641 | Yes |
| 91 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13289 | -0.828 | 0.0040 | Yes |