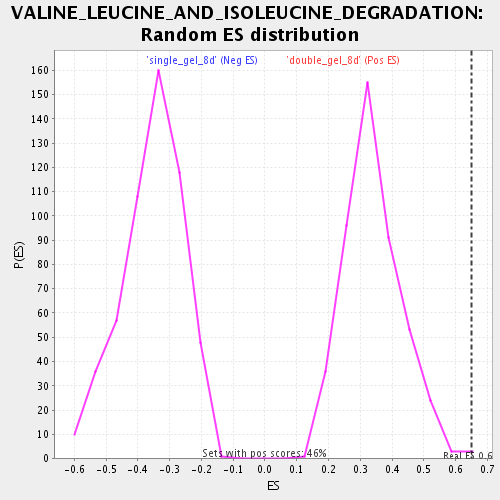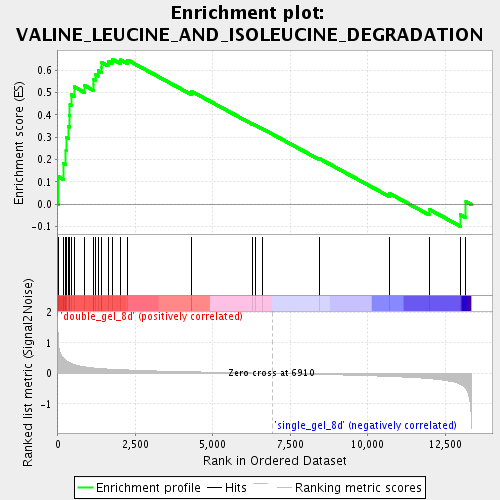
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_single_gel_8d.class.cls #double_gel_8d_versus_single_gel_8d_repos |
| Phenotype | class.cls#double_gel_8d_versus_single_gel_8d_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION |
| Enrichment Score (ES) | 0.648658 |
| Normalized Enrichment Score (NES) | 1.9030703 |
| Nominal p-value | 0.0021645022 |
| FDR q-value | 0.005292527 |
| FWER p-Value | 0.44 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 24 | 0.864 | 0.1232 | Yes |
| 2 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 172 | 0.499 | 0.1843 | Yes |
| 3 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 260 | 0.427 | 0.2395 | Yes |
| 4 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 271 | 0.419 | 0.2993 | Yes |
| 5 | HMGCL | HMGCL Entrez, Source | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria) | 344 | 0.371 | 0.3475 | Yes |
| 6 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 374 | 0.353 | 0.3964 | Yes |
| 7 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 389 | 0.344 | 0.4451 | Yes |
| 8 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 430 | 0.329 | 0.4897 | Yes |
| 9 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 524 | 0.292 | 0.5250 | Yes |
| 10 | MCCC1 | MCCC1 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | 856 | 0.213 | 0.5309 | Yes |
| 11 | HIBADH | HIBADH Entrez, Source | 3-hydroxyisobutyrate dehydrogenase | 1157 | 0.175 | 0.5337 | Yes |
| 12 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 1158 | 0.175 | 0.5590 | Yes |
| 13 | SDS | SDS Entrez, Source | serine dehydratase | 1222 | 0.169 | 0.5787 | Yes |
| 14 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 1296 | 0.164 | 0.5970 | Yes |
| 15 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 1399 | 0.157 | 0.6120 | Yes |
| 16 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 1403 | 0.157 | 0.6344 | Yes |
| 17 | PCCB | PCCB Entrez, Source | propionyl Coenzyme A carboxylase, beta polypeptide | 1620 | 0.142 | 0.6387 | Yes |
| 18 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 1747 | 0.134 | 0.6487 | Yes |
| 19 | ACADL | ACADL Entrez, Source | acyl-Coenzyme A dehydrogenase, long chain | 2024 | 0.121 | 0.6454 | No |
| 20 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 2248 | 0.113 | 0.6449 | No |
| 21 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 4305 | 0.053 | 0.4981 | No |
| 22 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4310 | 0.053 | 0.5054 | No |
| 23 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6279 | 0.012 | 0.3593 | No |
| 24 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 6381 | 0.010 | 0.3532 | No |
| 25 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 6602 | 0.006 | 0.3375 | No |
| 26 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 8449 | -0.033 | 0.2037 | No |
| 27 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 10696 | -0.098 | 0.0492 | No |
| 28 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 11983 | -0.171 | -0.0226 | No |
| 29 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 12985 | -0.357 | -0.0462 | No |
| 30 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13167 | -0.505 | 0.0131 | No |