|
Biorithm
1.1
|
|
Biorithm
1.1
|
Stores a set of points and information about the point's dimensions. This class is typically useful for storing one gene expression dataset. More...
#include <point.h>
Public Member Functions | |
| MyPointSet (const MyPointSet ©) | |
| const MyPointSet & | operator= (const MyPointSet &rhs) |
| void | compareTTest (string name, const MyPointSet &other, MyNT &tstat, MyNT &pvalue) const |
| Compare the expression values of point "name" in the invocant and in other by performing a t-test. | |
| MyNT | computeAverageExpressionValue (const set< string > &subset) |
| void | computeCoordinateDistribution (MyHistogram &histogram) const |
| Compute the distribution of coordinates in the point set. | |
| MyNT | computeMean (string point) const |
| Compute the mean value of the vectors corresponding to point. | |
| bool | computeMean (string point, MyPoint &mean) const |
| Compute the mean (vector) of the vectors corresponding to point. | |
| bool | computeMeanAndVariance (string point, MyNT &mu, MyNT &var, unsigned int &numCoords) const |
| void | computeMeanAndStandardDeviation (MyNT &mean, MyNT &stddev) |
| void | computeSignificantValues (MyNT pvalueThreshold, MyHistogram &histogram, MyNT &value) |
| void | convertToRanks () |
| Convert the coordinates of every point to their ranks in that point. | |
| void | print (string outfile) |
| Print the data set to a file. | |
| void | print (ostream &ostr) |
| Print the data set to an output stream. | |
| virtual void | read (MyClusterParams ¶ms, unsigned int whichPointSet=0) |
| virtual void | read (string infile, const MyAffyFileFormat &format=MyAffyFileFormat()) |
| virtual void | insert (const MyPoint &point) |
| virtual unsigned int | size () const |
| virtual unsigned int | getNumPoints () const |
| virtual unsigned int | getNumDimensions () const |
| virtual unsigned int | getNumSamples () const |
| virtual unsigned int | getNumGenes () const |
| virtual bool | isPoint (string name) const |
| virtual vector< unsigned int > | getPointIndex (string name) const |
| virtual const MyPoint & | getSample (unsigned int i) const |
| bool | doClassNamesExist () const |
| string | getClassName (int i) const |
| string | getSampleName (int i) const |
| string | getGeneName (int i) const |
| unsigned int | numInClass (string className) const |
| void | setClasses (string className) |
| void | setClasses (const MyClassesType &classes) |
| unsigned int | readClasses (string fileName) |
| void | getClasses (MyClassNamesType &names) |
| virtual const vector< MyDimInfo > & | getDimensionInfo () const |
| virtual MyDimInfo | getDimensionInfo (int i) const |
| virtual void | getDimensionNames (set< string > &names) const |
| Return the names of all the dimensions. | |
| virtual void | getDimCoords (int dim, vector< MyNT > &coords) const |
| virtual MyNT | getDimensionLow (int i) const |
| virtual MyNT | getDimensionHigh (int i) const |
| virtual MyNT | getDimensionMean (int i) const |
| virtual MyNT | getDimensionSigma (int i) const |
| virtual void | getDimensionValues (unsigned int i, vector< MyNT > &values) const |
| virtual unsigned int | getDimensionCount (unsigned int i, MyNT value) const |
| virtual const MyPoint & | operator[] (int i) const |
| virtual MyPoint & | operator[] (int i) |
| virtual void | getInterval (const vector< int > &discIndices, unsigned int dim, MyNT &low, MyNT &high) const |
| virtual string | suggestClass () const |
| virtual bool | generate (int &index, bool forbidChosen=true) const |
| virtual bool | generate (int num, vector< int > &indices, bool forbidHidden=true, string classToUse="") const |
| virtual int | getNumNotChosen () const |
| virtual int | getNumNotHidden () const |
| virtual void | getPointNames (set< string > &names) const |
| Return the names of all the points. | |
| virtual void | flip () |
| void | splitByClass (vector< MyPointSet > &splitPointSets, map< string, unsigned int > &classToIndex) |
| If there is more than one class for this point set, split the points by class. If there is just one class, return the point set. | |
| void | randomise () |
| virtual void | choosePoints (const vector< int > &containedPoints) |
| virtual void | hidePoints (const vector< int > &containedPoints) |
| virtual void | hide (const vector< int > &containedPoints, const vector< MyDimensionInterval > &intervals) |
| virtual MyNT | computeEntropy (int dim, MyVerbosityLevel verbosity) const |
| virtual void | computeEntropies (MyVerbosityLevel verbosity) |
| virtual unsigned int | filter (const MyClusterParams ¶ms) |
| unsigned int | filter (MyNT maxDownRegulated, MyNT minUpRegulated) |
| virtual bool | computeCorrelations (string point1, string point2, vector< MyNT > &correlations, string method="Pearsons") const |
| Compute the correlations between point1 and point2. | |
| virtual void | computeCorrelations (MyNT threshold, string id, bool absoluteValues=true, map< string, map< string, MyNT > > *pairs=NULL) |
| Based on the value of threshold, compute statistics on the correlations between all pairs of points and print them to a file whose name is based on id. | |
| virtual void | printCorrelations (MyNT threshold, string outputFile, bool absoluteValues=true, map< string, map< string, MyNT > > *pairs=NULL) |
| Compute the correlations between all pairs of points and print those greater than a threshold to a file. | |
| MyHistogram | printCorrelationsHistogram (string outputFile) const |
| Print a histogram of the correlations between all pairs of points to a file. | |
| MyHistogram | computeCorrelationsHistogram (map< string, map< string, MyNT > > *pairs=NULL) const |
| virtual MyHistogram | computeRandomisedCorrelationsHistogram (unsigned int numRandomisations, string outputFile) |
| void | standardise () |
| void | standardise (MyNT m, MyNT s) |
| void | storeRowNameTranslations (const map< string, set< string > > &rowNameAliases) |
| bool | _isFlipped () const |
Friends | |
| class | MyCluster |
Stores a set of points and information about the point's dimensions. This class is typically useful for storing one gene expression dataset.
| void MyPointSet::compareTTest | ( | string | name, |
| const MyPointSet & | other, | ||
| MyNT & | tstat, | ||
| MyNT & | pvalue | ||
| ) | const |
Compare the expression values of point "name" in the invocant and in other by performing a t-test.
| [in] | name,the | identifier of the point to be compared. |
| [in] | other,the | other instance of MyPointSet. |
| [out] | tstat,the | value of the t-statistic. |
| [out] | pvalue,the | p-value of the t-statistic. |
| MyNT MyPointSet::computeAverageExpressionValue | ( | const set< string > & | subset | ) |
Compute the average expression value of the points in subset.
The method computes the average expression value of all the points in subset over all the points in the data.
| bool MyPointSet::computeCorrelations | ( | string | point1, |
| string | point2, | ||
| vector< MyNT > & | correlations, | ||
| string | method = "Pearsons" |
||
| ) | const [virtual] |
Compute the correlations between point1 and point2.
Compute the correlation between two points.
| [in] | point1,the | ID of the first point. |
| [in] | point2,the | ID of the second point. |
| [out] | correlations,a | vector of computed correlation values. |
| [in] | method,the | type of correlation to compute. This option can take the following values: |
Pearsons: compute the Pearson's correlation coefficient.
pearsons: same as Pearsons.
discrete: compute the discrete correlation coefficient. See the documentation of MyPoint::computeDiscreteCorrelation() for an explanation.
This method takes the IDs of two points. For every pair of indices corresponding to the points (multiple points may have the same ID), the method computes the correlation between that pair of indices.
| [in] | point1,name | of the first point. |
| [in] | point2,name | of the second point. |
| [out] | correlations,a | vector of correlation values. |
Given the names point1 and point2 of two points in the invocant, compute the correlations between all pairs of points with these names.
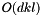 , where
, where 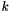 is the number of points and
is the number of points and 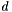 is the number of dimensions in the data. It can take a long time!
is the number of dimensions in the data. It can take a long time!| void MyPointSet::computeCorrelations | ( | MyNT | threshold, |
| string | id, | ||
| bool | absoluteValues = true, |
||
| map< string, map< string, MyNT > > * | pairs = NULL |
||
| ) | [virtual] |
Based on the value of threshold, compute statistics on the correlations between all pairs of points and print them to a file whose name is based on id.
| [in] | ::MyNT | threshold, only print correlations greater than this parameter. |
| [in] | ::string | id, a string to construct the name of the file to print the correlations to. If this string is the empty string, the method does not print any correlations. |
| [in] | absoluteValues; | if this boolean is true (the default value), the method computes absolute values of the correlations. |
| [out] | pairs,a | pointer to a 2-dimensional map. If the pointer is not NULL, then the method stores all the pairwise correlations in this map. The method ignores this argument unless the absolute value of thresh is at most 1. |
| MyHistogram MyPointSet::computeCorrelationsHistogram | ( | map< string, map< string, MyNT > > * | pairs = NULL | ) | const |
Compute a histogram of the correlations between all pairs of points.
| [out] | pairs,a | pointer to a 2-dimensional map. If the pointer is not NULL, then the method stores all the pairwise correlations in this map. |
| void MyPointSet::computeMeanAndStandardDeviation | ( | MyNT & | mean, |
| MyNT & | stddev | ||
| ) |
Compute the mean and standard deviation of the distribution of coordinates in the point set.
| bool MyPointSet::computeMeanAndVariance | ( | string | point, |
| MyNT & | mu, | ||
| MyNT & | var, | ||
| unsigned int & | numCoords | ||
| ) | const |
Compute the mean and variance of the coordinates of the average of the vectors corresponding to point.
| [in] | point,the | name of the point. |
| [out] | mu,the | required mean. |
| [out] | variance,the | required variance. |
| [out] | numCoords,the | number of values used to calculate the mean and standard deviation. |
| MyHistogram MyPointSet::computeRandomisedCorrelationsHistogram | ( | unsigned int | numRandomisations, |
| string | outputFile | ||
| ) | [virtual] |
Randomises the set of points multiple times, computes the correlations between all pairs of points for each random set, and computes a histogram of all the correlations.
Randomises the set of points multiple times, computes the correlations between all pairs of points for each random set, and computes a histogram of all the correlations.
| unsigned | int numRandomisations, the number of times to randomise the data. |
| string | outputFile, the stub of the name of the file to output the histogram to. The default value of this argument is "". The method adds the string "-randomised-num-trials" to outputFile where "num" is the value of numRandomisations. If the file name is provided, the method prints each histogram to the file. A blank line separates two histograms. |
| virtual bool MyPointSet::isPoint | ( | string | name | ) | const [inline, virtual] |
Check if the point name is a point in the invocant.
| [in] | name,the | name of the point. |
| void MyPointSet::printCorrelations | ( | MyNT | threshold, |
| string | outputFile, | ||
| bool | absoluteValues = true, |
||
| map< string, map< string, MyNT > > * | pairs = NULL |
||
| ) | [virtual] |
Compute the correlations between all pairs of points and print those greater than a threshold to a file.
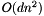 , where
, where 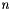 is the number of points and
is the number of points and 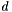 is the number of dimensions in the data. It can take a long time!
is the number of dimensions in the data. It can take a long time!| [in] | ::MyNT | threshold, only print correlations greater than this parameter. |
| [in] | ::string | outputFile, the name of the file to print the correlations to. If this string is the empty string, the method does not print any correlations. The method adds the string "-correlations-threshold-val", where val is the string representation of the parameter "threshold" to the string outputFile. |
| [in] | absoluteValues; | if this boolean is true (the default value), the method computes absolute values of the correlations. |
| [out] | pairs,a | pointer to a 2-dimensional map. If the pointer is not NULL, then the method stores all the pairwise correlations in this map. The method ignores this argument unless the absolute value of thresh is at most 1. |
| MyHistogram MyPointSet::printCorrelationsHistogram | ( | string | outputFile | ) | const |
Print a histogram of the correlations between all pairs of points to a file.
| string | outputFile, the name of the file to print the histogram to. The method adds the string "-correlations-histogram" to the name of this file before printing. If the file already exists, the method simply reads the histogram from that file. |
| void MyPointSet::randomise | ( | ) | [inline] |
Constructs a random permutation of the coordinates of each point.
| void MyPointSet::splitByClass | ( | vector< MyPointSet > & | splitPointSets, |
| map< string, unsigned int > & | classToIndex | ||
| ) |
If there is more than one class for this point set, split the points by class. If there is just one class, return the point set.
| [out] | splitPointSets,a | vector containing the split set. |
| [out] | classToIndex,a | map from class name to indices in the splitPointSets vector. |
friend class MyCluster [friend] |
 1.7.6.1
1.7.6.1