|
Biorithm
1.1
|
|
Biorithm
1.1
|
An algorithm where one label/function competes against an army of all the other labels/functions. More...
#include <gain-algorithm.h>
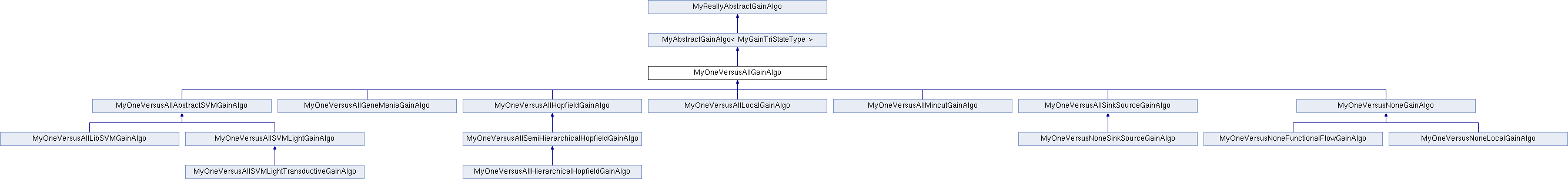
Public Member Functions | |
| MyOneVersusAllGainAlgo (MyGainGraph &g, MyAnnotations &a, MyGainParams &p, Reporter &r, GeneOntology *go=NULL) | |
| virtual void | computeCrossValidationResults (const BioFunction &function, const MyNodeIdList &cvNodes) |
| Compute results of cross validation. | |
| virtual void | computeCut (MyNodeIdList &nodesToAnnotate, MyNodeIdSet &cutNodes) |
| virtual MyNT | computeEnergy (const MyNodeIdList &nodesToAnnotate) |
| virtual void | computePredictionConfidences (const BioFunction &function, MyNodeIdList &nodesToAnnotate, MyNodeIdList &predictedNodes) |
| virtual void | computePredictions (const BioFunction &function, const MyNodeIdList &nodesToAnnotate, MyNodeIdList &predictedNodes) |
| virtual MyGainTriState | computeState (MyNode &node) |
| virtual void | constructCrossValidationLists (vector< MyNodeIdList > &cvLists) |
| Construct lists of nodes for cross validation. | |
| virtual bool | hasConverged (const MyNodeIdList &nodesToAnnotate) |
| Check if the states of the nodes in the argument nodes have converged. | |
| virtual void | maskNodeStates (const MyNodeIdList &nodes) |
| virtual string | getName () |
| virtual void | initialiseNodeStates (const BioFunction &function, MyNodeIdList &nodesToAnnotate) |
| virtual void | run (const MyNodeIdList &nodesToAnnotate) |
| virtual void | visualisePredictions (const BioFunction ¤tFunction, MyNodeIdList &nodesToAnnotate) |
| virtual void | visualiseCut (const BioFunction ¤tFunction, MyNodeIdList &predictedNodes) |
| Layout a graph that highlights the cut seprating nodes annotated with or predicted to be annotated with currentFunction from all the other nodes. | |
| virtual void | printStatistics (ostream &statsStream) |
| virtual void | initialiseEdgeWeightsCutoff (const BioFunction &function) |
| virtual void | initialiseEdgeWeightsDepth (const BioFunction &function) |
Protected Attributes | |
| bool | _allowApproximateConvergence |
| map< BioFunction, MyNT > | _finalEnergies |
| map< BioFunction, MyNT > | _initialEnergies |
| map< BioFunction, unsigned int > | _numIterations |
| map< BioFunction, unsigned int > | _numOperations |
| unsigned int | _numTruePathViolatingAnnotations |
| unsigned int | _numTruePathViolatingPredictions |
| map< string, set< string > > | _truePathViolatingAnnotations |
| map< string, set< string > > | _truePathViolatingPredictions |
An algorithm where one label/function competes against an army of all the other labels/functions.
This algorithm computes node states as the weighted average of the states of the neighbours of a node. In effect, it solves a linear system whose variables are the node states.
| void MyOneVersusAllGainAlgo::computeCrossValidationResults | ( | const BioFunction & | function, |
| const MyNodeIdList & | cvNodes | ||
| ) | [virtual] |
Compute results of cross validation.
After invoking crossValidate(), compute the results. Each algorithm needs to define this method, since the algorithm stores the node states.
Implements MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllGeneManiaGainAlgo, and MyOneVersusAllSinkSourceGainAlgo.
| void MyOneVersusAllGainAlgo::computeCut | ( | MyNodeIdList & | nodesToAnnotate, |
| MyNodeIdSet & | cutNodes | ||
| ) | [virtual] |
Compute which genes in nodesToAnnotate MyOneVersusNoneShortestPathGainAlgo::run() predicts has the function.
| nodesToAnnotate,an | instance of MyNodeIdList, the nodes that run() tried to annotate. |
| predictedNodes,an | instance of MyNodeIdList, the nodes that run() predicts as having the function. |
Reimplemented from MyAbstractGainAlgo< MyGainTriStateType >.
| MyNT MyOneVersusAllGainAlgo::computeEnergy | ( | const MyNodeIdList & | nodesToAnnotate | ) | [virtual] |
Compute the energy of the FLN.
The energy of the FLN is the sum of the inconsistencies of the edges in the FLN. The inconsistency of an edge is the product of the weight of the edge and the states of the nodes incident on the edge. If the edge weight is positive, the more similar the states are, the more consistent the edge is and the smaller the energy is. The same is true when the edge weight is negative and the states are different.
| void MyOneVersusAllGainAlgo::computePredictions | ( | const BioFunction & | function, |
| const MyNodeIdList & | nodesToAnnotate, | ||
| MyNodeIdList & | predictedNodes | ||
| ) | [virtual] |
Compute which genes in nodesToAnnotate MyOneVersusAllGainAlgo::run() predicts has the function. Since different algorithms may have different methods for predicting a function, this method must be implemented separately for each class.
| nodesToAnnotate,an | instance of MyNodeIdList, the nodes that run() tried to annotate. |
| predictedNodes,an | instance of MyNodeIdList, the nodes that run() predicts as having the function. |
Implements MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllGeneManiaGainAlgo, MyOneVersusNoneFunctionalFlowGainAlgo, MyOneVersusAllSinkSourceGainAlgo, and MyOneVersusAllSemiHierarchicalHopfieldGainAlgo.
| MyGainTriState MyOneVersusAllGainAlgo::computeState | ( | MyNode & | node | ) | [virtual] |
This method should be a member of this class since the method needs access to the states of the nodes (stored in subclasses of the class).
Implements MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllGeneManiaGainAlgo, MyOneVersusNoneLocalGainAlgo, MyOneVersusNoneFunctionalFlowGainAlgo, MyOneVersusAllSinkSourceGainAlgo, MyOneVersusAllHopfieldGainAlgo, and MyOneVersusAllLocalGainAlgo.
| void MyOneVersusAllGainAlgo::constructCrossValidationLists | ( | vector< MyNodeIdList > & | cvLists | ) | [virtual] |
Construct lists of nodes for cross validation.
| [out] | cvLists,a | vector of MyNodeIdLists. |
Each element of this vector contains a list of node ids (stored in a MyNodeIdList) whose annotation details should be deleted for cross validation. In the case of leave-one-out cross validation, each element of the vector will contain just one node.
Implements MyAbstractGainAlgo< MyGainTriStateType >.
| virtual string MyOneVersusAllGainAlgo::getName | ( | ) | [inline, virtual] |
Return the name of the algorithm.
Implements MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllGeneManiaGainAlgo, MyOneVersusNoneLocalGainAlgo, MyOneVersusNoneFunctionalFlowGainAlgo, MyOneVersusNoneSinkSourceGainAlgo, MyOneVersusAllSinkSourceGainAlgo, MyOneVersusAllSVMLightTransductiveGainAlgo, MyOneVersusAllSVMLightGainAlgo, MyOneVersusAllLibSVMGainAlgo, MyOneVersusAllHierarchicalHopfieldGainAlgo, MyOneVersusAllSemiHierarchicalHopfieldGainAlgo, MyOneVersusAllMincutGainAlgo, MyOneVersusAllHopfieldGainAlgo, MyOneVersusAllLocalGainAlgo, and MyOneVersusNoneGainAlgo.
| bool MyOneVersusAllGainAlgo::hasConverged | ( | const MyNodeIdList & | nodesToAnnotate | ) | [virtual] |
Check if the states of the nodes in the argument nodes have converged.
Implements MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllGeneManiaGainAlgo, and MyOneVersusAllSinkSourceGainAlgo.
| void MyOneVersusAllGainAlgo::initialiseNodeStates | ( | const BioFunction & | function, |
| MyNodeIdList & | nodesToAnnotate | ||
| ) | [virtual] |
Initialise the states of the nodes in the graph. Each node annotated with function gets the state ANNOTATED_STATE; these nodes are "clamped," i.e., the algorithm will not change their states. All other nodes get the state HYPOTHETICAL_STATE. The algorithm may change the states of these nodes. In other words, these nodes are not clamped. The argument nodesToAnnotate contains the IDs of the unclamped nodes.
For every node ID in nodes, "remember" its state information and clamp it, so that MyOneVersusNoneGainAlgo::run() can't modify the state of that node.
| [in] | nodes,the | list of node whose initial state must be unmasked. |
Implements MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllGeneManiaGainAlgo, MyOneVersusNoneSinkSourceGainAlgo, MyOneVersusAllSinkSourceGainAlgo, MyOneVersusAllSemiHierarchicalHopfieldGainAlgo, and MyOneVersusNoneGainAlgo.
| void MyOneVersusAllGainAlgo::maskNodeStates | ( | const MyNodeIdList & | nodes | ) | [virtual] |
For every node ID in nodes, "forget" its state information and unclamp it, so that MyOneVersusNoneGainAlgo::run() can modify the state of that node.
| [in] | nodes,the | list of node whose initial state must be masked. |
Implements MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllGeneManiaGainAlgo, and MyOneVersusAllSinkSourceGainAlgo.
| void MyOneVersusAllGainAlgo::printStatistics | ( | ostream & | statsStream | ) | [virtual] |
Print statistics about the performance of the algorithm.
For this algorithm, this method simply prints the number of iterations taken to achieve convergence.
Reimplemented from MyReallyAbstractGainAlgo.
Reimplemented in MyOneVersusAllSemiHierarchicalHopfieldGainAlgo.
| void MyOneVersusAllGainAlgo::run | ( | const MyNodeIdList & | nodesToAnnotate | ) | [virtual] |
This method repeatedly iterates over all the nodes in nodesToAnnotate in the order in which they occur in nodesToAnnotate. It computes the state of each node in this list. The method iterates until the states converge.
Implements MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllGeneManiaGainAlgo, MyOneVersusNoneLocalGainAlgo, MyOneVersusNoneFunctionalFlowGainAlgo, MyOneVersusAllSinkSourceGainAlgo, MyOneVersusAllAbstractSVMGainAlgo, MyOneVersusAllMincutGainAlgo, MyOneVersusAllHopfieldGainAlgo, and MyOneVersusAllLocalGainAlgo.
| void MyOneVersusAllGainAlgo::visualiseCut | ( | const BioFunction & | currentFunction, |
| MyNodeIdList & | predictedNodes | ||
| ) | [virtual] |
Layout a graph that highlights the cut seprating nodes annotated with or predicted to be annotated with currentFunction from all the other nodes.
The method lays out the graph induced by the edges in the cut and all the nodes on the "currentFunction" side of the cut.
Reimplemented from MyAbstractGainAlgo< MyGainTriStateType >.
Reimplemented in MyOneVersusAllAbstractSVMGainAlgo.
 1.7.6.1
1.7.6.1